Chromatin control of environmental stress response
Dr Ritwick Sawarkar
We aim to understand the different ways in which our cells respond to chemicals from the external environment such as the drugs we take. We hope to identify how we can avoid side-effects of drugs especially RNA therapeutics like anti-sense oligos and mRNA vaccines.
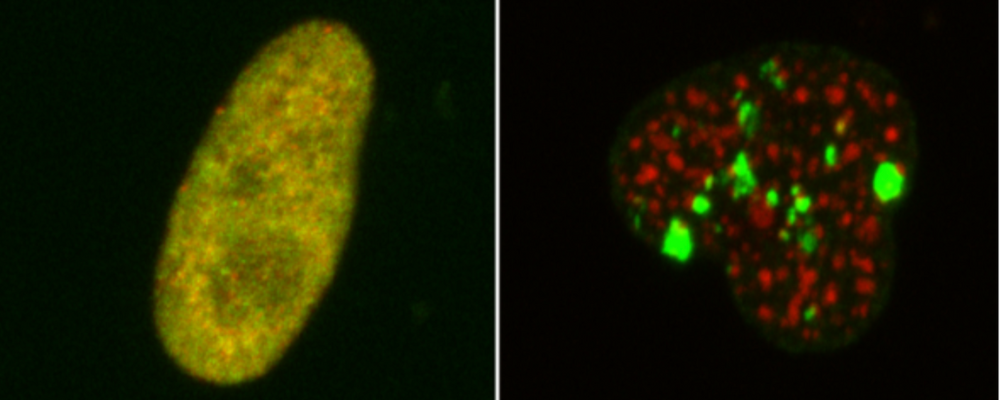
Cells respond to environmental stress and toxins by mounting an adaptive stress response in order to survive stressful conditions. Transcriptional control is a major regulatory layer that determines the strength, the duration and persistence of cellular stress response. Transcription factors, chromatin modifications and non-coding RNA influence the transcriptional response to environmental stress. The molecular mechanisms by which chromatin exerts control over stress response is the main focus of the Unit programme. We aim to address the following questions:
- Which cellular pathways sense environmental stress/ toxins and signal to the genome?
- How does chromatin interpret the information about cellular health and toxic exposure determining the transcriptional response to stress?
- How does the transcriptional response adapt cellular phenotypes to survive the stress?
We study these three questions in the context of cellular exposure to environmental stress as well as small-molecule therapeutics in collaboration with pharmaceutical companies. Our approaches include genomics, single-cell transcriptomics, proteomics, chromatin biochemistry as well as genome-wide screening to identify novel components of stress-response pathways. Discovery-driven global approaches in mammalian cells are further validated by in vitro reconstitution experiments and mouse genetic models. We aim to gain novel insights and mechanistic understanding of transcriptional response to stress and toxins.
Latest Sawarkar group news
Key publications
Leone S, Srivastava A, Herrero-Ruiz A, Hummel B, Tittel L, Campalastri R, Aprile-Garcia F, Tan JH, Rawat P, Andersson P, Willis WE, Sawarkar R. HSP70 binds to specific non-coding RNA and regulates human RNA polymerase III. Mol Cell. 84(4), 687-701, (2024).
Rawat P, Boehning M, Hummel B, Aprile-Garcia F, Pandit AS, Eisenhardt N, Khavaran A, Niskanen E, Vos SM, Palvimo JJ, Pichler A, Cramer P, Sawarkar R. Stress-induced nuclear condensation of NELF drives transcriptional downregulation. Mol Cell. 81 (5), 1013-1026, (2021)
Antonova A, Hummel B, Khavaran A, Redhaber D, Aprile Garcia F, Rawat P, Gundel K, Schneck M, Hansen E, Mitschke J, Mittler G, Miething C and Sawarkar R. Heat-shock protein 90 controls the expression of cell-cycle genes by stabilizing metazoan-specific Host-Cell Factor HCFC1. Cell Reports. 29(6):1645-1659. (2019)
Aprile-Garcia F, Tomar P, Hummel B, Khavaran A and Sawarkar R. Nascent-protein ubiquitination is required for heat shock-induced gene downregulation in human cells. Nat Struct Mol Biol. 26(2):137-146. (2019)
Hummel B, Hansen EC, Yoveva A, Aprile-Garcia F, Hussong R and Sawarkar R. The evolutionary capacitor HSP90 buffers the regulatory effects of mammalian endogenous retroviruses. Nat Struct Mol Biol. 24, 234-242. (2017)
Sawarkar R, Sievers C and Paro R. Hsp90 globally targets paused RNA polymerase to regulate gene expression in response to stimuli. Cell. 149(4):807-18. (2012)